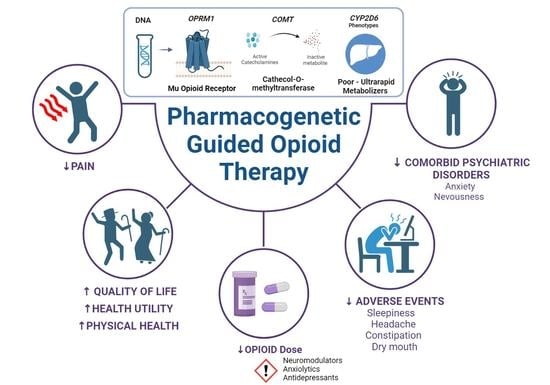
Pharmacogenetic Guided Opioid Therapy Improves Chronic Pain Outcomes and Comorbid Mental Health: A Randomized, Double-Blind, Controlled Study
Abstract
:1. Introduction
2. Results
2.1. Patient Demographic and Genetic Data
2.2. Pain Management Efficacy
2.3. Pain Management Tolerability
3. Discussion
Limitations
4. Materials and Methods
4.1. Study Design and Participants
4.2. Data Collection and Opioid Titration
4.2.1. Clinical Outcomes
4.2.2. Pharmacology and Safety Data
4.3. Genetic Determination
4.4. Recommendations Based on Pharmacogenetics
4.5. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goesling, J.; Lin, L.A.; Clauw, D.J. Psychiatry and Pain Management: At the Intersection of Chronic Pain and Mental Health. Curr. Psychiatry Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Owusu Obeng, A.; Hamadeh, I.; Smith, M. Review of Opioid Pharmacogenetics and Considerations for Pain Management. Pharmacotherapy 2017, 37, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Bright, D.; Petry, N.; Roath, E.; Reckow, E.; Chavour, S. Barriers, solutions, and effect of using pharmacogenomics data to support opioid prescribing. J. Manag. Care Spec. Pharm. 2020, 26, 1597–1602. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Callaghan, J.T.; Samer, C.F.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Klein, T.E.; Caudle, K.E.; Haidar, C.E.; Shen, D.D.; Callaghan, J.T.; Sadhasivam, S.; et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450 2D6 Genotype and Codeine Therapy: 2014 Update. Clin. Pharmacol. Ther. 2014, 95, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Gaedigk, A.; Sangkuhl, K.; Whirl-Carrillo, M.; Klein, T.; Steven Leeder, J. Prediction of CYP2D6 Phenotype from Genotype across World Populations. Genet. Med. 2017, 19, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Zahari, Z.; Ismail, R. Influence of Cytochrome P450, Family 2, Subfamily D, Polypeptide 6 (CYP2D6) Polymorphisms on Pain Sensitivity and Clinical Response to Weak Opioid Analgesics. Drug Metab. Pharmacokinet. 2014, 29, 29–43. [Google Scholar] [CrossRef] [Green Version]
- Eckhardt, K.; Li, S.; Ammon, S.; Schänzle, G.; Mikus, G.; Eichelbaum, M. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain 1998, 76, 27–33. [Google Scholar] [CrossRef]
- Lötsch, J.; Rohrbacher, M.; Schmidt, H.; Doehring, A.; Brockmöller, J.; Geisslinger, G. Can Extremely Low or High Morphine Formation from Codeine Be Predicted Prior to Therapy Initiation? Pain 2009, 144, 119–124. [Google Scholar] [CrossRef]
- Poulsen, L.; Arendt-Nielsen, L.; Brøsen, K.; Sindrup, S.H. The Hypoalgesic Effect of Tramadol in Relation to CYP2D6*. Clin. Pharmacol. Ther. 1996, 60, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Skarke, C.; Schmidt, H.; Rohrbacher, M.; Hofmann, U.; Schwab, M.; Geisslinger, G. Evidence for Morphine-Independent Central Nervous Opioid Effects after Administration of Codeine: Contribution of Other Codeine Metabolites. Clin. Pharmacol. Ther. 2006, 79, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.; Laforge, K.S.; Tian, M.; Melia, D.; Zhang, S.; Borg, L.; Gong, J.; Schluger, J.; Strong, J.A.; Leal, S.M.; et al. Single-Nucleotide Polymorphism in the Human Mu Opioid Receptor Gene Alters β-Endorphin Binding and Activity: Possible Implications for Opiate Addiction. Proc. Natl. Acad. Sci. USA 1998, 95, 9608–9613. [Google Scholar] [CrossRef] [Green Version]
- Candiotti, K.A.; Yang, Z.; Rodriguez, Y.; Crescimone, A.; Sanchez, G.C.; Takacs, P.; Medina, C.; Zhang, Y.; Liu, H.; Gitlin, M.C. The Impact of CYP2D6 Genetic Polymorphisms on Postoperative Morphine Consumption. Pain Med. 2009, 10, 799–805. [Google Scholar] [CrossRef]
- Lopes, G.S.; Bielinski, S.; Moyer, A.M.; Jacobson, D.J.; Wang, L.; Jiang, R.; Larson, N.B.; Miller, V.M.; Zhu, Y.; Cavanaugh, D.C.; et al. Sex Differences in Type and Occurrence of Adverse Reactions to Opioid Analgesics: A Retrospective Cohort Study. BMJ Open 2021, 11, 44157. [Google Scholar] [CrossRef]
- Liu, S.; Kang, W.J.; Abrimian, A.; Xu, J.; Cartegni, L.; Majumdar, S.; Hesketh, P.; Bekker, A.; Pan, Y.X. Alternative Pre-Mrna Splicing of the Mu Opioid Receptor Gene, Oprm1: Insight into Complex Mu Opioid Actions. Biomolecules 2021, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Fredrikson, K.M.; Fasolino, T. Pharmacogenetic Testing: Clinical Integration and Application for Chronic Pain Management. Nurse Pract. 2021, 46, 12–19. [Google Scholar] [CrossRef]
- Lötsch, J.; Geisslinger, G. Are μ-Opioid Receptor Polymorphisms Important for Clinical Opioid Therapy? Trends Mol. Med. 2005, 11, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Craddock, N.; Owen, M.J.; O’Donovan, M.C. The Catechol-O-Methyl Transferase (COMT) Gene as a Candidate for Psychiatric Phenotypes: Evidence and Lessons. Mol. Psychiatry 2006, 11, 446–458. [Google Scholar] [CrossRef] [Green Version]
- Zubieta, J.K.; Heitzeg, M.M.; Smith, Y.R.; Bueller, J.; Xu, K.; Xu, Y.; Koeppe, R.; Stohler, C.; Goldman, D. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science 2003, 299, 1240–1243. [Google Scholar] [CrossRef]
- Kambur, O.; Männistö, P.T. Catechol-O-Methyltransferase and Pain. Int. Rev. Neurobiol. 2010, 95, 227–279. [Google Scholar] [CrossRef] [PubMed]
- Aygun Kocabas, N. Catechol-O-Methyltransferase (COMT) Pharmacogenetics in the Treatment Response Phenotypes of Major Depressive Disorder (MDD). CNS Neurol. Disord. Drug Targets 2012, 11, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Tunbridge, E.M.; Harrison, P.J. Importance of the COMT Gene for Sex Differences in Brain Function and Predisposition to Psychiatric Disorders. Curr. Top Behav. Neurosci. 2011, 8, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Ira, E.; Zanoni, M.; Ruggeri, M.; Dazzan, P.; Tosato, S. COMT, Neuropsychological Function and Brain Structure in Schizophrenia: A Systematic Review and Neurobiological Interpretation. J. Psychiatry Neurosci. 2013, 38, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.D.; Parvataneni, H.K.; Gray, C.F.; Deen, J.T.; Prieto, H.A.; Pulido, L.F.; Elsey, A.R.; Elwood, E.N.; Starostik, P.; Gong, Y.; et al. A Hybrid Implementation-Effectiveness Randomized Trial of CYP2D6 Guided Post-Operative Pain Management. Genet. Med. 2021, 23, 621. [Google Scholar] [CrossRef]
- Jarvis, J.P.; Peter, A.P.; Keogh, M.; Baldasare, V.; Beanland, G.M.; Wilkerson, Z.T.; Kradel, S.; Shaman, J.A. Real-World Impact of a Pharmacogenomics-Enriched Comprehensive Medication Management Program. J. Pers. Med. 2022, 12, 421. [Google Scholar] [CrossRef]
- Swen, J.J.; van der Wouden, C.H.; Manson, L.E.; Abdullah-Koolmees, H.; Blagec, K.; Blagus, T.; Böhringer, S.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.C.; et al. A 12-Gene Pharmacogenetic Panel to Prevent Adverse Drug Reactions: An Open-Label, Multicentre, Controlled, Cluster-Randomised Crossover Implementation Study. Lancet 2023, 401, 347–356. [Google Scholar] [CrossRef]
- Linares, O.A.; Daly, D.; Linares, A.D.; Stefanovski, D.; Boston, R.C. Personalized Oxycodone Dosing: Using Pharmacogenetic Testing and Clinical Pharmacokinetics to Reduce Toxicity Risk and Increase Effectiveness. Pain Med. 2014, 15, 791–806. [Google Scholar] [CrossRef]
- Orliaguet, G.; Hamza, J.; Couloigner, V.; Denoyelle, F.; Loriot, M.A.; Broly, F.; Garabedian, E.N. A Case of Respiratory Depression in a Child with Ultrarapid CYP2D6 Metabolism After Tramadol. Pediatrics 2015, 135, e753–e755. [Google Scholar] [CrossRef] [Green Version]
- Elkalioubie, A.; Allorge, D.; Robriquet, L.; Wiart, J.F.; Garat, A.; Broly, F.; Fourrier, F. Near-Fatal Tramadol Cardiotoxicity in a CYP2D6 Ultrarapid Metabolizer. Eur. J. Clin. Pharmacol. 2011, 67, 855–858. [Google Scholar] [CrossRef] [Green Version]
- MacHado-Alba, J.E.; Serna-Echeverri, L.S.; Valladales-Restrepo, L.F.; MacHado-Duque, M.E.; Gaviria-Mendoza, A. Use of Tramadol or Other Analgesics in Patients Treated in the Emergency Department as a Risk Factor for Opioid Use. Pain Res. Manag. 2020, 2020, 8847777. [Google Scholar] [CrossRef] [PubMed]
- Schelde, A.B.; Sørensen, A.M.S.; Hindsø, M.; Christensen, M.B.; Jimenez-Solem, E.; Eriksson, R. Sex and age differences among tramadol users in three Nordic countries. Dan. Med. J. 2020, 67, A06190336. [Google Scholar] [PubMed]
- Barrachina, J.; Margarit, C.; Muriel, J.; López-Gil, V.; López-Gil, S.; Ballester, P.; Mira-Lorente, L.; Agulló, L.; Peiró, A.M. Sex Differences in Oxycodone/Naloxone vs. Tapentadol in Chronic Non-Cancer Pain: An Observational Real-World Study. Biomedicines 2022, 10, 2468. [Google Scholar] [CrossRef]
- Patel, V.; Saxena, S.; Lund, C.; Thornicroft, G.; Baingana, F.; Bolton, P.; Chisholm, D.; Collins, P.Y.; Cooper, J.L.; Eaton, J.; et al. The Lancet Commission on Global Mental Health and Sustainable Development. Lancet 2018, 392, 1553–1598. [Google Scholar] [CrossRef] [Green Version]
- Ostovar-Kermani, T.; Arnaud, D.; Almaguer, A.; Garcia, I.; Gonzalez, S.; Mendez Martinez, Y.H.; Surani, S. Painful Sleep: Insomnia in Patients with Chronic Pain Syndrome and Its Consequences. Folia Med. 2020, 62, 645–654. [Google Scholar] [CrossRef]
- Yadav, D.; Askew, R.L.; Palermo, T.; Li, L.; Andersen, D.K.; Chen, M.; Fisher, W.E.; Fogel, E.L.; Forsmark, C.E.; Hart, P.A.; et al. Association of Chronic Pancreatitis Pain Features with Physical, Mental, and Social Health. Clin. Gastroenterol. Hepatol. 2022, 21, 1781–1791.e4. [Google Scholar] [CrossRef] [PubMed]
- Patten, R.K.; Tacey, A.; Bourke, M.; Smith, C.; Pascoe, M.; Vogrin, S.; Parker, A.; McKenna, M.J.; Tran, P.; De Gori, M.; et al. The Impact of Waiting Time for Orthopaedic Consultation on Pain Levels in Individuals with Osteoarthritis: A Systematic Review and Meta-Analysis. Osteoarthr. Cartil. 2022, 30, 1561–1574. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Walsh, D.; Brito-Dellan, N. Opioid and adjuvant analgesics: Compared and contrasted. Am. J. Hosp. Palliat. Care 2011, 28, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.; Muriel, J.; Peiró, A.M. CYP2D6 Phenotypes and Opioid Metabolism: The Path to Personalized Analgesia. Expert Opin. Drug Metab. Toxicol. 2022, 18, 261–275. [Google Scholar] [CrossRef]
- Garnæs, K.K.; Mørkved, S.; Tønne, T.; Furan, L.; Vasseljen, O.; Johannessen, H.H. Mental health among patients with chronic musculoskeletal pain and its relation to number of pain sites and pain intensity, a cross-sectional study among primary health care patients. BMC Musculoskelet Disord. 2022, 23, 1115. [Google Scholar] [CrossRef]
- Dorado, P.; González, I.; Naranjo, M.E.; de Andrés, F.; Peñas-Lledó, E.M.; Calzadilla, L.R.; LLerena, A. Lessons from Cuba for Global Precision Medicine: CYP2D6 Genotype Is not a Robust Predictor of CYP2D6 Ultrarapid Metabolism. OMICS 2017, 21, 17–26. [Google Scholar] [CrossRef]
- Ruiz-Cantero, M.T.; Blasco-Blasco, M.; Chilet-Rosell, E.; Peiró, A.M. Gender Bias in Therapeutic Effort: From Research to Health Care. Farm. Hosp. 2020, 44, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, G.; Prkachin, K.M.; Kaseweter, K.A.; Williams, A.C.D.C. Health Care Providers’ Judgments in Chronic Pain: The Influence of Gender and Trustworthiness. Pain 2016, 157, 1618–1625. [Google Scholar] [CrossRef] [Green Version]
- Barrachina, J.; Muriel, J.; Margarit, C.; Planelles, B.; Ballester, P.; Richart-Martínez, M.; Cutillas, E.; Zandonai, T.; Morales, D.; Peiró, A.M. Global Pain State Questionnaire: Reliability, Validity, and Gender Gap. Arch. Intern. Med. Res. 2021, 4, 91–113. [Google Scholar] [CrossRef]
- Ware, J., Jr.; Kosinski, M.; Keller, S.D. A 12-Item Short-Form Health Survey: Construction of scales and preliminary tests of reliability and validity. Med. Care 1996, 34, 220–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snaith, R.P. Health and Quality of Life Outcomes the Hospital Anxiety and Depression Scale. Health Qual. Life Outcomes 2003, 1, 29. [Google Scholar] [CrossRef] [Green Version]
- Pergolizzi, J.; Böger, R.H.; Budd, K.; Dahan, A.; Erdine, S.; Hans, G.; Kress, H.G.; Langford, R.; Likar, R.; Raffa, R.B.; et al. Opioids and the Management of Chronic Severe Pain in the Elderly: Consensus Statement of an International Expert Panel with Focus on the Six Clinically Most Often Used World Health Organization Step III Opioids (Buprenorphine, Fentanyl, Hydromorphone, Methadone, Morphine, Oxycodone). Pain Pract. 2008, 8, 287–313. [Google Scholar] [CrossRef]
- Gaedigk, A.; Simon, S.D.; Pearce, R.E.; Bradford, L.D.; Kennedy, M.J.; Leeder, J.S. The CYP2D6 Activity Score: Translating Genotype Information into a Qualitative Measure of Phenotype. Clin. Pharmacol. Ther. 2008, 83, 234–242. [Google Scholar] [CrossRef]
- Margarit, C.; Roca, R.; Inda, M.d.M.; Muriel, J.; Ballester, P.; Moreu, R.; Conte, A.L.; Nuñez, A.; Morales, D.; Peiró, A.M. Genetic Contribution in Low Back Pain: A Prospective Genetic Association Study. Pain Pract. 2019, 19, 836–847. [Google Scholar] [CrossRef]
- Smith, D.M.; Weitzel, K.W.; Elsey, A.R.; Langaee, T.; Gong, Y.; Wake, D.T.; Duong, B.Q.; Hagen, M.; Harle, C.A.; Mercado, E.; et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: A pragmatic clinical trial. Genet Med. 2019, 21, 1842–1850. [Google Scholar] [CrossRef]
- Dong, H.; Lu, S.J.; Zhang, R.; Liu, D.D.; Zhao, Y.Z.; Song, C.Y. Effect of the CYP2D6 gene polymorphism on postoperative analgesia of tramadol in Han nationality nephrectomy patients. Eur. J. Clin. Pharmacol. 2015, 71, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Bastami, S.; Haage, P.; Kronstrand, R.; Kugelberg, F.C.; Zackrisson, A.-L.; Uppugunduri, S. Pharmacogenetic aspects of tramadol pharmacokinetics and pharmacodynamics after a single oral dose. Forensic Sci. Int. 2014, 238, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Lane, K.; Dixon, J.J.; McKeown, D.; Johnston, A.; Van Schaik, R.H.N.; Van Fessem, M.; Macphee, I.A.M.; Philips, B.J. Using tramadol to measure CYP2D6 metabolism in critically ill adults. Intensive Care Med. 2014, 40, 1177–1178. [Google Scholar] [CrossRef] [PubMed]
- Haage, P.; Kronstrand, R.; Josefsson, M.; Calistri, S.; van Schaik, R.H.N.; Green, H.; Kugelberg, F.C. Enantioselective pharmacokinetics of tramadol and its three main metabolites; impact of CYP2D6, CYP2B6, and CYP3A4 genotype. Pharmacol. Res. Perspect. 2018, 6, e00419. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Naito, T.; Sato, H.; Hiraide, T.; Yamada, Y.; Kawakami, J. Impact of CYP genotype and inflammatory markers on the plasma concentrations of tramadol and its demethylated metabolites and drug tolerability in cancer patients. Eur. J. Clin. Pharmacol. 2018, 74, 1461–1469. [Google Scholar] [CrossRef]
- Arafa, M.H.; Atteia, H.H. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6) are associated with long term tramadol treatment-induced oxidative damage and hepatotoxicity. Toxicol. Appl. Pharmacol. 2018, 346, 37–44. [Google Scholar] [CrossRef]
- Yu, H.; Hong, S.; Jeong, C.H.; Bae, J.W.; Lee, S. Development of a linear dual column HPLCMS/ MS method and clinical genetic evaluation for tramadol and its phase I and II metabolites in oral fluid. Arch. Pharm. Res. 2018, 41, 288–298. [Google Scholar] [CrossRef]
- Yu, H.; Choi, M.; Jang, J.-H.; Park, B.; Seo, Y.H.; Jeong, C.-H.; Bae, J.-W.; Lee, S. Development of a column-switching LC-MS/MS method of tramadol and its metabolites in hair and application to a pharmacogenetic study. Arch. Pharm. Res. 2018, 41, 554–563. [Google Scholar] [CrossRef]
- Fonseca, S.; Amorim, A.; Costa, H.A.; Franco, J.; Porto, M.J.; Santos, J.C.; Dias, M. Sequencing CYP2D6 for the detection of poor-metabolizers in post-mortem blood samples with tramadol. Forensic Sci. Int. 2016, 265, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Batistaki, C.; Chrona, E.; Kostroglou, A.; Kostopanagiotou, G.; Gazouli, M. CYP2D6 Basic Genotyping of Patients with Chronic Pain Receiving Tramadol or Codeine. A Study in a Greek Cohort. Pain Med. 2020, 21, 3199–3204. [Google Scholar] [CrossRef]
- Dagostino, C.; Allegri, M.; Napolioni, V.; D’Agnelli, S.; Bignami, E.; Mutti, A.; van Schaik, R.H. CYP2D6 genotype can help to predict effectiveness and safety during opioid treatment for chronic low back pain: Results from a retrospective study in an Italian cohort. Pharmgenomics Pers. Med. 2018, 11, 179–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, T.; Takashina, Y.; Yamamoto, K.; Tashiro, M.; Ohnishi, K.; Kagawa, Y.; Kawakami, J. CYP3A5*3 affects plasma disposition of noroxycodone and dose escalation in cancer patients receiving oxycodone. J. Clin. Pharmacol. 2011, 51, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Stamer, U.M.; Zhang, L.; Book, M.; Lehmann, L.E.; Stuber, F.; Musshoff, F. CYP2D6 genotype dependent oxycodone metabolism in postoperative patients. PLoS ONE 2013, 8, e60239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreassen, T.N.; Eftedal, I.; Klepstad, P.; Davies, A.; Bjordal, K.; Lundström, S.; Kaasa, S.; Dale, O. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A crosssectional multicentre study. Eur. J. Clin. Pharmacol. 2012, 68, 55–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balyan, R.; Mecoli, M.; Venkatasubramanian, R.; Chidambaran, V.; Kamos, N.; Clay, S.; Moore, D.L.; Mavi, J.; Glover, C.D.; Szmuk, P.; et al. CYP2D6 pharmacogenetic and oxycodone pharmacokinetic association study in pediatric surgical patients. Pharmacogenomics 2017, 18, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Slanar, O.; Dupal, P.; Matouskova, O.; Vondrackova, H.; Pafko, P.; Perlik, F. Tramadol efficacy in patients with postoperative pain in relation to CYP2D6 and MDR1 polymorphisms. Bratislavske Lekarske Listy 2012, 113, 152–155. [Google Scholar] [CrossRef] [Green Version]



| CNCP Patients | Total (n = 50) | PGx-Guided (n = 28) | Usual Care (n = 22) | |
|---|---|---|---|---|
| Women % (n) | 80 (40) | 93 (26) * | 64 (14) | |
| Age (years old, mean (SD)) | 59 (14) | 59 (14) | 60 (14) | |
| Employment status % (n) | ||||
| Active | 32 (16) | 32 (9) | 32 (7) | |
| Disability | 22 (11) | 21 (6) | 23 (5) | |
| Retired | 20 (10) | 21 (6) | 18 (4) | |
| Homemaker | 18 (9) | 14 (4) | 23 (5) | |
| Unemployment | 8 (4) | 11 (3) | 5 (1) | |
| Genetic data % (n) | ||||
| CYP2D6 | PMs | 4 (2) | 4 (1) | 5 (1) |
| IMs | 42 (21) | 43 (12) | 41 (9) | |
| EMs | 48 (24) | 46 (13) | 50 (11) | |
| UMs | 6 (3) | 7 (2) | 5 (1) | |
| OPRM1 | AA | 62 (31) | 68 (19) | 55 (12) |
| AG | 30 (15) | 28 (8) | 32 (7) | |
| Mutant-GG | 8 (4) | 4 (1) | 14 (3) | |
| COMT | GG | 12 (6) | 14 (4) | 9 (2) |
| GA | 52 (26) | 39 (11) | 68 (15) | |
| Mutant-AA | 36 (18) | 46 (15) | 23 (5) | |
| Baseline | Final (3 Months) | |||
|---|---|---|---|---|
| Mean (SD), median [IQR] or % | PGx-guided n = 28 | Usual care n = 22 | PGx-guided n = 28 | Usual care n = 22 |
| Pain intensity (0–100 mm) | 76 (21)×× | 71 (26) | 59 (20) | 80 (19) ** |
| Pain relief (0–100 mm) | 28 (27) | 35 (32) | 48 (29) × | 35 (31) |
| Quality of life (0–100 mm) | 43 (21) | 47 (22) | 56 (15) ** | 38 (22) |
| Health Utility (0–1 scores) | 0.30 [0.06–0.65] | 0.20 [0.03–0.65] | 0.71 [0.58–0.82] * × | 0.51 [0.13–0.67] |
| HAD-Anxiety (0–21 sco.) | 8 [5–11] × | 7 [5–10] | 5 [3–7] | 9 [5–12] |
| HAD-Depression (0–21 sco.) | 7 [4–9] | 8 [4–12] | 4 [3–7] | 8 [2–12] |
| Physical (SF12) | 27 (7) | 28 (7) | 36 (8) ×× | 29 (9) |
| Mental (SF12) | 43 (14) | 41 (12) | 43 (7) | 43 (12) |
| MEDD (mg/day) | 34 [23–56] | 49 [36–112] | 35 [22–61] | 60 [40–80] * |
| Simple analgesics | 68 * | 36 | 71 | 64 |
| NSAIDs | 21 | 9 | 21 | 14 |
| Tramadol | 39 | 27 | 61 | 50 |
| Fentanyl | 0 | 9 | 0 | 18 * |
| Oxycodone | 11 | 5 | 4 | 9 |
| Tapentadol | 0 | 5 | 21× | 9 |
| Buprenorphine | 7 | 5 | 7 | 0 |
| Neuromodulators | 50 | 32 | 57 | 64 |
| Antidepressants | 36 | 41 | 39 | 50 |
| Anxiolytics | 43 | 45 | 32 | 45 |
| Baseline | Final (3 Months) | |||
|---|---|---|---|---|
| Median [IQR]) or % | PGx-guided n = 28 | Usual care n = 22 | PGx-guided n = 28 | Usual care n = 22 |
| Total Adverse Events | 3 [1–5] ×× | 4 [3–6] | 1 [0–2] | 4 [2–6] ** |
| Insomnia | 29 | 59 *× | 7 | 23 |
| Sleepiness | 25× | 32 | 0 | 23 * |
| Depression | 25× | 18 | 0 | 5 |
| Nervousness | 29 | 45 | 11 | 41 * |
| Dry mouth | 25 | 55 * | 18 | 50 * |
| Headache | 25 | 36 | 11 | 45 ** |
| Constipation | 21 | 32 | 14 | 41 * |
| Weight change | 25 | 27 | 4 | 14 |
| Dizziness | 21 | 32 | 11 | 18 |
| Itching | 18 | 27 | 11 | 23 |
| Loss of libido | 11 | 14 | 7 | 5 |
| Dry Skin | 14 | 14 | 11 | 32 |
| Nausea | 7 | 23 | 7 | 5 |
| Lack of appetite | 7 | 18 | 7 | 18 |
| Edema | 4 | 5 | 0 | 5 |
| Redness of skin | 0 | 5 | 0 | 5 |
| Sexual disturbance | 0 | 9 | 0 | 9 |
| Vomiting | 0 | 14 | 0 | 5 |
| Opioids | Tramadol Codeine | Oxycodone Hydrocodone | Tapentadol Buprenorphine Fentanyl |
|---|---|---|---|
| CYP2D6 Phenotye | |||
| UMs | Avoid codeine and tramadol use because of serious toxicity potential | Use of oxycodone with caution/vigilance. Increase surveillance for increased adverse events | No recommendation |
| NMs | Use tramadol label recommended age-specific or weight-specific dosing | No recommendation | No recommendation |
| IMs | Use tramadol label recommended age-specific or weight-specific dosing | No recommendation | No recommendation |
| PMs | Avoid codeine and tramadol use because of the possible decrease in analgesic response | Use of oxycodone/hydrocodone with caution/vigilance | No recommendation |
| OPRM1 Genotype | |||
| AA | The presence of the AA wild-type genotype implies a lower dose required to achieve analgesia | ||
| AG/GG | The presence of the G allele implies a higher dose required to achieve analgesia | ||
| COMT Genotype | |||
| AG/AA | The presence of the A allele implies a lower dose required to achieve analgesia | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agulló, L.; Aguado, I.; Muriel, J.; Margarit, C.; Gómez, A.; Escorial, M.; Sánchez, A.; Fernández, A.; Peiró, A.M. Pharmacogenetic Guided Opioid Therapy Improves Chronic Pain Outcomes and Comorbid Mental Health: A Randomized, Double-Blind, Controlled Study. Int. J. Mol. Sci. 2023, 24, 10754. https://doi.org/10.3390/ijms241310754
Agulló L, Aguado I, Muriel J, Margarit C, Gómez A, Escorial M, Sánchez A, Fernández A, Peiró AM. Pharmacogenetic Guided Opioid Therapy Improves Chronic Pain Outcomes and Comorbid Mental Health: A Randomized, Double-Blind, Controlled Study. International Journal of Molecular Sciences. 2023; 24(13):10754. https://doi.org/10.3390/ijms241310754
Chicago/Turabian StyleAgulló, Laura, Isidro Aguado, Javier Muriel, César Margarit, Alba Gómez, Mónica Escorial, Astrid Sánchez, Alicia Fernández, and Ana M. Peiró. 2023. "Pharmacogenetic Guided Opioid Therapy Improves Chronic Pain Outcomes and Comorbid Mental Health: A Randomized, Double-Blind, Controlled Study" International Journal of Molecular Sciences 24, no. 13: 10754. https://doi.org/10.3390/ijms241310754